The present state
The OSE system consists of the following modules:
BioImage
image processing, detection of phenotypes
Intended purpose of the software is a general image processing framework. it has an open and modular architecture where the user can easily write custom algorithms, bundle them into a module and plug into BioImage without modifying the BioImage source code. The software is open source and will be available soon.
Created in MPI-CBG
KNIME
A modular data exploration platform
Created at Konstanz University
TracePilot
Visualizes objects over time in 2D or 3D

Created in BIOTEC
Cell
Detective
3D cell tracking
The software aim is to track dividing cells in 3D environment. Images originate from confocal microscopy. The software is under development.
Created in MPI-CBG
Dynomica
Epidetect
Predicting the immune response
Antibodies are raised by the immune system against regions on the surface of a protein known as epitopes. An epitope, or antigenic determinant, is defined as a region of 6-12 amino acid or carbohydrate residues to which an antibody can bind. A continuous epitope is composed of a contiguous stretch of residues in a protein sequence. In order to predict or design a B-cell epitope, various properties from the protein sequence, are calculated including secondary structure, hydrophilicity, surface probability, flexibility, antigenic index, amphiphilicity. For this reason many algorithms were implemented and integrated into one software solution.
BioImageXD
Analysis, processing and 3D rendering of multi dimensional microscopy images
BioImageXD is a collaborative open source free software project, designed and developed by microscopists, cell biologists and programmers from the Universities of Jyväskylä and Turku in Finland, Max Planck Institute CBG, Dresden, Germany and collaborators worldwide.
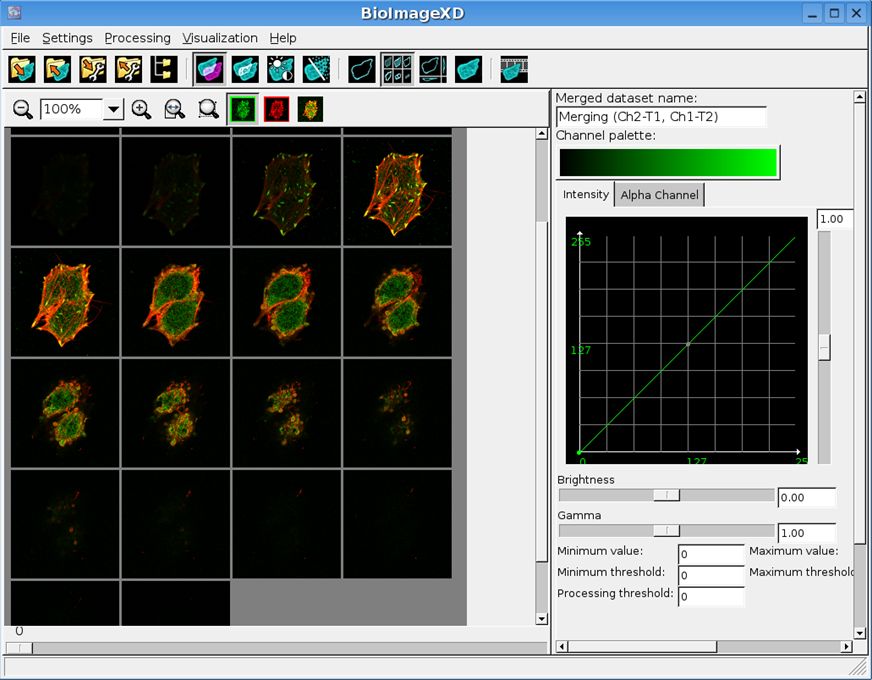
Created by BioImageXD